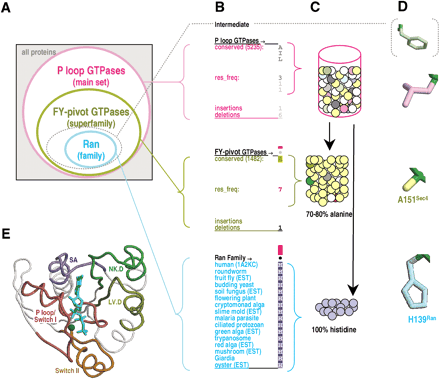
CHAIN analysis representational schemes. The examples shown inA–D correspond to the hierarchical alignment in Figure 1. (A) Venn diagram representing hierarchical relationships between aligned sequence sets. The dotted oval corresponds to a hypothetical intermediate category consisting of conserved residues in Ran that fall outside the categories of this particular hierarchy. (B) Notation used within hierarchical alignments. Position 139 of Ran is shown. The main and superfamily sets, which contain too many sequences to display directly, are represented in the alignment as position-specific conserved patterns. (The total number of sequences for these two categories is shown in parentheses.) The corresponding residue frequencies (‘res_freq’) are given in integer tenths below conserved residues. For example, a ‘5’ in integer tenths indicates that the corresponding residue directly above it occurs in 50%–60% of the (weighted) sequences. Insertion and deletion frequencies are similarly given in integer tenths (black; range 10%–100%) or hundredths (gray; range 1%–9%) as indicated. Ran family aligned residues are displayed directly. Histogram bar heights are approximately logarithmically proportional to the measure of selective constraint (see Methods), as defined by the following urn model. (C) Urn model for measuring the selective constraint acting on a specific position. The residues observed in the main set at this position are modeled as distinctly colored balls in an urn. Some of the colors are similar (representing biochemically similar amino acids). The selective constraint is then defined as the difficulty of drawing by chance at least as many of the same- or similarly-colored balls from the urn as are observed in the subalignments (in this case, alanine for the FY-pivot superfamily or histidine for the Ran family). Note that our analysis of the Ran family uses the main set as the ‘superalignment’ urn (see Methods); alternatively, the FY-pivot GTPases may also be used as the superalignment urn, though the sparser data set would yield less accurate background frequency estimates. Note that the alignments in Figures 1A and 1B measure sequence constraints using a standard background model (see Methods). (D) Color scheme used for residue side-chains in Figures 4 F5 F6 F7 F8 F9. (E) Color scheme for structural regions described in the text and figures. The structure of Sec4p (pdb code: 1G17) is shown in complex with a GTP analog (cyan) and magnesium (dark green).











