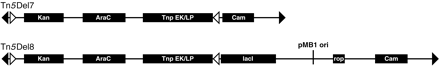
Structure and features of transposons used in this work. The structure of Tn5Del7 and Tn5Del8 is conceptually the same. Both transposons are defined by IE sequences CTGTCTCTTGATCAGATCT (open triangle indicates IE sequence). Two ME sequences CTGTCTCTTATACACATCT (filled triangle indicates ME sequence) are faced toward each other, defining donor DNA for transposition using these ends. Distance between the tips of the IE and ME ends on the left is 64 bp. This is the size of the linker that remains in the chromosome after deletion. Donor DNA encodes the Tnp gene for ME end-mediated transposition under the control of an arabinose inducible promoter and also a KmRgene (Kan). Between the right pair of transposon ends, both transposons have a selectable marker (Cam). The only difference is that Tn5Del8 has a conditional origin of replication. Lac repressor encoded by the lacI gene controls the origin. Moderate plasmid (after its formation) copy number is ensured by Rop function.











