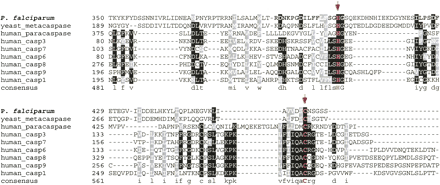
Multiple alignment of the catalytic regions of the putative P. falciparum metacaspase (PF13_0289) and the representative proteolytically active caspases using Clustal X1.8 followed by manual correction. The catalytic dyad H (404) and C (460), which are part of the active sites, is highlighted with arrowheads. Graphic presentation of the alignment and the consensus sequence were obtained by the program BOXSHADE 3.21. Conserved residues are shaded with black and gray. The accession numbers of caspase protein sequences used for alignment refer to Figure 3.











