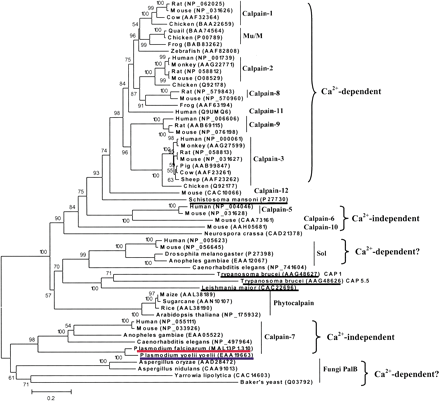
The phylogenetic tree of the calpains, inferred by the neighbor-joining method based on the amino acid sequences with Poisson corrected distance. The option of complete deletion of gaps was used for tree construction. 1000 bootstrap replicates were used to infer the reliability of branching points. Bootstrap values of >50% are presented. The scale bar indicates the number of amino acid substitutions per site. The parasite sequences are underlined. The putative P. falciparum calpain and P. yoelii yoeliiortholog are highlighted in red and blue, respectively. The accession number for each sequence is included in the parentheses after the species name.











