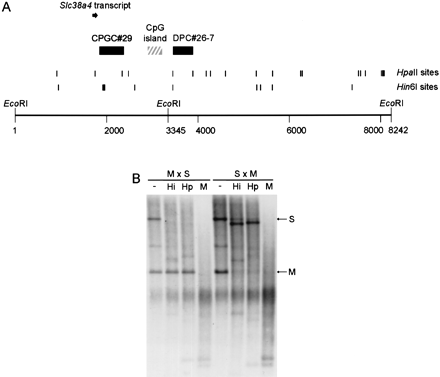
Identification of Slc38a4 and methylation analysis of the locus. (A) Map of locus indicating relative locations of Me-RDA clones (solid boxes) to a CpG island (hatched box) and the first exon of Slc38a4 (solid arrow). The ATG translation start forSlc38a4 is located in exon 2; downstream exons are not illustrated in this figure. The EcoRI sites indicated are those used in methylation analysis; the site at position 3345 is polymorphic between M. musculus and M. spretus. The RT-PCR assay used to analyze imprinted expression was located in the body of the gene and is not shown here. Hin6I andHpaII sites are given as vertical lines; in some instances, individual sites could not be resolved. (B) Methylation analysis in adult livers from crosses between M. musculus(C57BL/6, M) female and M. spretus (S) male (M × S, maternal inheritance of M alleles, paternal inheritance of S allele), and from backcross animals ([C57BL/6 × M. spretus] × C57BL/6) that inherit a maternal M. spretusallele and a paternal M. musculus allele (S × M) at this locus. The DNA was digested with EcoRI alone (−) or in combination with Hin6I (Hi), HpaII (Hp), orMspI (M).











