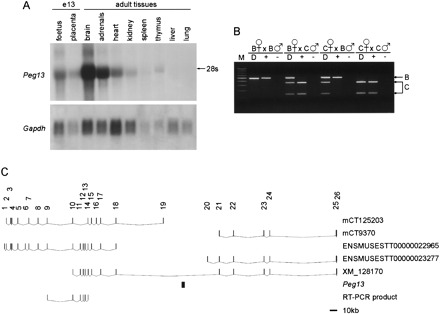
Tissue-specific and imprinted expression of Peg13. (A) Northern blot hybridization of total RNA from adult tissues and midgestation (e13) fetus and placenta, using Peg13as a probe. The size of the transcript on the Northern blot is, in comparison to 18S/28S rRNA, consistent with the full-length transcript sequence. Hybridization with Gapdh gives an indication of the amount of RNA loaded. (B) RT-PCR/RFLP analysis of imprinted expression of Peg13 in inter-subspecific hybrids of C57BL/6 (B) and CAST/Ei (C) using an RT-PCR product from within the sequence of EST AW125295. KspI digests the PCR product from the CAST/Ei allele and assigns the parental origin of alleles in RT-PCRs (+). Control RT reactions with no reverse transcriptase were included (−). As the transcript runs contiguous to the DNA, PCR amplification from genomic DNA (D) indicates the presence of both alleles in the hybrid embryos. M is a 100-bp molecular weight marker. (C) Map indicating the position of Peg13 within a large intron of the gene encoding the mouse homolog of the predicted protein KIAA1882. mCT125203 and mCT9370 are Celera transcripts; ENSMUSEST00000022965 and ENSMUSEST00000023277 are Ensembl entries. XM_128170 indicates that the KIAA1882 gene spans the region containing the DPC#19–2 transcript; both genes are transcribed in the same transcriptional orientation. The RT-PCR product used to investigate imprinting of mouse KIAA1882 is shown.











