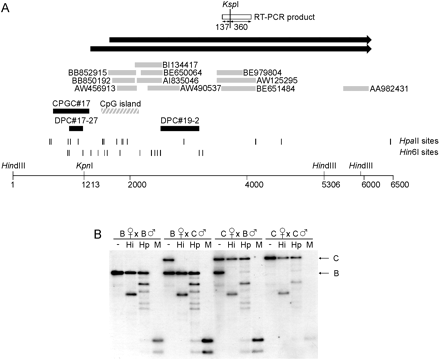
Identification and methylation analysis of the locus containingPeg13. (A) Map of locus containing Hin6I Me-RDA products DPC#19–2 and DPC#17–27 and CPGC#17 (solid boxes). The region contains a CpG island, indicated by a hatched box. Sequence matches to ESTs are indicated by gray boxes. The black arrows indicate the paternally expressed noncoding transcript, Peg13. Peg13 is contiguous with the genomic DNA and was identified in two isoforms, with lengths 4419 nt and 4723 nt, which differ at their 5′ ends. The HindIII sites flank the region analyzed by Southern blot hybridization; the KpnI site is polymorphic between C57BL/6 and CAST/Ei and specifically digests the C57BL/6 allele. The RT-PCR product used for expression analysis by using aKspI RFLP is shown as a white box; the restriction fragment lengths are given. Hin6I and HpaII sites are given as vertical lines; in some instances, individual sites could not be resolved. (B) Methylation analysis in day-13 fetuses from reciprocal crosses (B × C and C × B) between C57BL/6 (B) and CAST/Ei (C). The DNA was digested with HindIII andKpnI (−) or in combination with Hin6I (Hi),HpaII (Hp), or MspI (M). DNA from fetuses of the parental strain are included as controls.











