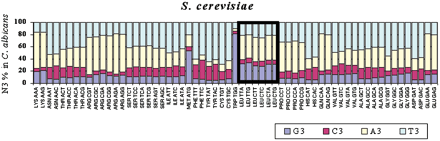
GC pressure alone at the third codon position does not explain the evolution of leucine codons in Candida albicans. To elucidate the role of GC pressure in the evolution of C. albicans ORFs, an analysis of GC3 pressure (GC pressure at the third codon position) was carried out by comparing the complete ORFs set of both genomes using BLASTP at an E-value of 10−5. For eachSaccharomyces cerevisiae codon, the corresponding N3 codon position was identified in C. albicans ortholog genes. The data show that S. cerevisiae codons are represented in C. albicans mainly by A- or T-terminating codons (yellow and light blue bars), in agreement with high AT pressure at the third codon position in the C. albicans genome (N3 = 71% AT; Table 2). However, the leucine codons show a small deviation from the pattern observed for all other codons. That is, they change more frequently than expected into G-terminating codons (blue bar), thus contradicting the high AT pressure at the N3 position observed in C. albicans genes. This increase in G3 is matched by a slight increase in A3, indicating an increase in purines at N3. This is achieved by decreasing the usage frequency of C- and T-ending codons. That is, when compared with all other codons, there is a relative increase in purines at the N3 position in C. albicans genes instead of an increase in AT-ending codons, as would be expected from the relative increase of AT pressure at N3 in the C. albicans genome, thus indicating that other forces apart from GC pressure shaped the evolution of leucine codons in the latter.











