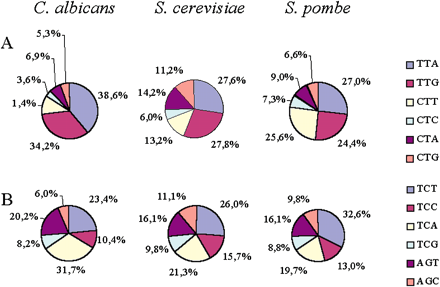
Usage of leucine and serine codons in Candida albicans,Saccharomyces cerevisiae, and Schizosaccharomyces pombe. (A) Of the six leucine codons, TTA and TTG are the most frequently used by the nuclear genomes of the three yeast species, the exception being CTT in S. pombe. In C. albicans, the usage of CTN codons, in particular CTC and CTA but also CTG codons, is repressed in relation to the same codons in the other two yeasts, whereas usage of the CTT codon is similar between C. albicansand S. cerevisiae. The usage of the CTA codon is also repressed in C. albicans, indicating that other forces apart from genome AT pressure shape CTN usage in this species. (B) The bias in C. albicans CTN usage is not observed for serine codons, whose distribution follows the expected pattern for a genome with low GC content in coding sequences (Table 2). The values indicated are relative to the total synonymous codon count for each genome and independent of amino acid frequency.











