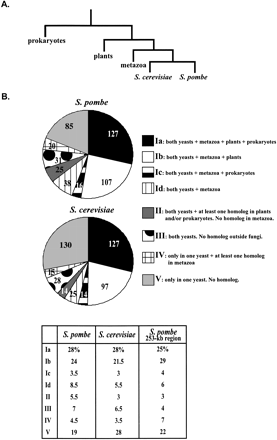
Comparison of S. pombe and S. cerevisiae proteome. (A) A consensus phylogeny of fission yeast and budding yeast adapted from Sipiczki (2000). (B) 450 proteins from S. pombe and S. cerevisiae (YPD and PombePD) were compared to proteins from prokaryotes, metazoa, plants, S. pombe, andS. cerevisiae using BLASTP (Altschul and Lipman 1990) with a cutoff E value of 10−5. The proteins from both yeasts were classified into eight different classes, according to the distribution of homologous proteins in other species. Class I: homologous proteins are found in both S. pombe and S. cerevisiae and in either metazoa + plants + prokaryotes (Ia), or metazoa + plants with no homolog in prokaryotes (Ib), or metazoa + prokaryotes (Ic), or only metazoa (Id). Class II: homologous proteins are found in both yeasts and in plants or prokaryotes, but there is no homolog in metazoa. Class III: homologous proteins are present in both S. cerevisiae and S. pombe, but there is no homolog outside the fungal branch. Class IV: homologous proteins of S. pombe are not found in S. cerevisae and S. cerevisiae proteins do not have an homolog in S. pombe, but homologous proteins of both yeasts are found at least in the metazoa branch. Class V: there is no homologous protein of one yeast in the other yeast, and no homolog in other branches. Numbers of genes in each category are shown for both yeasts. Percentages are given in the table. The third column gives the gene distribution in the 253-kb region that we selected for systematic gene deletion in this study.











