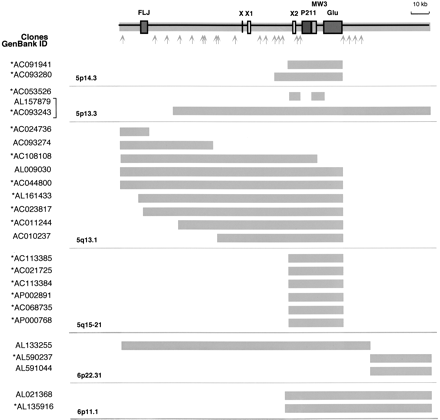
Paralogy map and sequence content of the HSA5/HSA6 duplicon. The genomic organization of the duplicated region was deduced from theAL157879 and AL133255 clones that were chosen as reference seed sequence. The thick gray lines illustrate the homology extent between paralogous loci. The chromosomal localization and the GeneBank ID of the clones are indicated. The P211, FLJ, and GUSB gene-derived sequences are boxed in dark gray. For exon-intron organization of theGUSB-derived sequences, see Figure 3A. The THE-1 transposable element MW3 is boxed in light gray. X, X1, and X2 are sequences paralogous to exons of the chimeric genes described in Figure 6. Gray arrows indicate the localization of sequences highly homologous (>95%) to ESTs and human UNIGENE clusters as follows: Hs.186379, Hs.312136, Hs.224604, Hs.145839, Hs.274528, Hs.297663, Hs.50454, Hs.202243, Hs.183256, Hs.132586, AW963166, BE780832, Hs.317160, Hs.326016, Hs.121081, Hs.294040, Hs.166361, Hs.321499, Hs.324135, Hs.7569, Hs.131950, and Hs.186180. The brackets indicate that the same clone was sequenced twice. (*) Clones in HTGS phase in GenBank—according to the June, 2002 freeze of the genome draft sequence.











