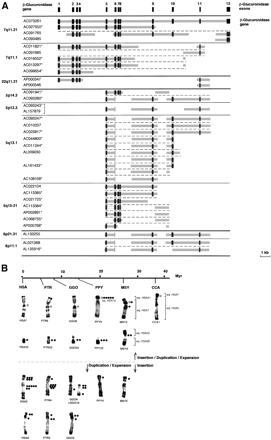
(A) Schematic representation of the genomic structure of the β-glucuronidase gene (GUSB) and GUSB-derived paralogous sequences. The genomic structure of the GUSB gene was established according to sequence comparison and alignment between the β-glucuronidase mRNA complete sequence (M15182) and genomic sequence of the BAC clone RP11-252P18 (AC073261). Dark gray boxes indicate the exons, and thick light-gray lines indicate intervening intronic sequences. For each GUSB-derived sequence found in the draft sequence of the human genome, the chromosomal localization and the GeneBank ID of the clone(s) they were derived from are indicated. (*) Clones in HTGS phase in GenBank—according to the June 2002 freeze of the genome draft sequence. Horizontal black bold lines illustrate chromosomal separations, horizontal black plain lines, subchromosomal separations, and horizontal black broken lines, gene-copy separations. The bracket indicates that the same clone was sequenced twice. (B) Summary of the in situ hybridization results obtained with a Glu 5–10 cosmid probe on human (HSA) and other primate chromosomes. A molecular timescale for primate evolution is indicated. The number of dots indicates the average signal intensity observed at a given location, resulting from the number of loci and the hybridization efficiency. The brackets illustrate the homologies between Macaca or Cebus and human chromosomes. In the presumed ancestral karyotype of placental mammals, the HSA7 homolog was composed of two parts. These two components fused before the separation of Cercopithecoidea and Hominidae (∼25 Mya), and then pericentric and paracentric inversions occurred in theHominidae lineage. Thus, HSA7 is a fairly recent chromosome shared by HSA and PTR only. In Cebus capucinus, the smallest part homolog to HSA7 is on CCA1 associated with the homolog to the whole HSA5 (Richard et al. 1996). In Macaca sylvana, MSY2 is formed by the equivalents of the whole HSA7 and HSA21, and MSY9 by the equivalents of HSA20 and HSA22 (Muleris et al. 1984). HSA22/PTR23/GGO23/PPY23 on one hand, and HSA6/PTR5/GGO5 on the other hand, differ mostly only by heterochromatic variations (Yunis and Prakash 1982). The differences in the chronology of expansion/spreading of the GUSB paralogous sequences is depicted by a horizontal broken line, top, primo-insertion in pericentromeric regions on ancestral HSA7 and HSA22, bottom, primo-insertion in the ancestral HSA5p euchromatic region.











