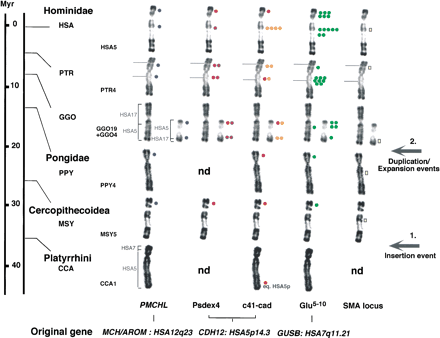
Summary of the in situ hybridization results of the gene-derived sequences/SMA genes on human and other primate chromosomes equivalent to human chromosome 5 (HSA5). The scale indicates the generally accepted times of divergence of Anthropoids from the human lineage (Hacia 2001). On the basis of the phylogenetic classification ofGoodman (1999), we placed Pongo in the familyPongidae and grouped Gorilla, Pan, andHomo in the family Hominidae. The same color code as in Figure 1 was used. The number of dots indicates the average signal intensity observed at a given location with a particular clone. Red dots on Psdex4 and c41-cad hybridization patterns indicate cross-hybridization with the genuine CDH12 gene. The names and chromosomal localizations of the original genes, whose gene segments derived from are indicated at bottom. The yellow bar indicates hybrization signals obtained with two genes (SMNand NAIP) specific to the SMA locus. The position of the homologs to human SMA locus is indicated as a chromosomal index of the region equivalent to HSA 5q13. Brackets indicate the homology between HSA and other primate chromosomes. For Cebus capucinus (CCA), CCA1 is formed by the equivalent of the whole HSA5, plus a small segment of HSA7 (Richard et al. 1996). For Macaca sylvana(MSY) and Pongo pygmaeus (PPY), the equivalent to HSA5 (MSY5 and PPY4, respectively) have a banding pattern very similar to HSA5 (Dutrillaux 1979). For Gorilla gorilla (GGO), GGO4 and GGO19 are the products of a reciprocal translocation between ancestral chromosomes equivalent to HSA5 and HSA17, respectively (Dutrillaux et al. 1973). Evolutionary breakpoints occurred in regions equivalent to HSA5q13.3 and HSA17p12 (Stankiewicz et al. 2001). Brackets indicate the homology between GGO and HSA chromosomes. For Pan troglodytes(PTR), PTR4 differs from HSA5 by a pericentromeric inversion, inv(5)(p14.3;q13.3) (Yunis and Prakash 1982; Marzella et al. 1997). Grey lines on PTR chromosomes indicate the pericentromeric inversion breakpoints.











