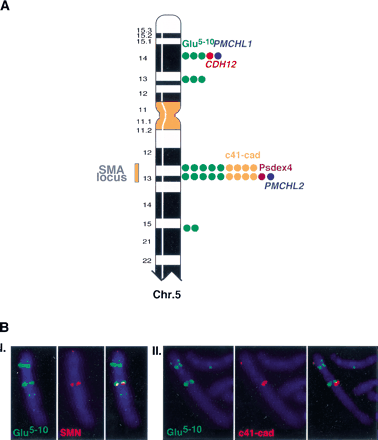
(A) Colocalization of the gene-derived sequences on human chromosome 5. The FISH hybrization signals observed on human chromosome 5 with gene segment-specific probes are represented by colored dots. (Blue) PMCHL genes; (green) Glu 5–10; (red)Br-cadherin gene (CDH12); (orange) c41-cad; (purple) Psdex4. The number of dots is proportional to the signal intensity, which was scored on a scale of from 0–5. The yellow bar delimits the predisposition locus to the SMA disease. (B) Illustration of some dual-color FISH experiments. (I.) Glu 5–10 biotin-labeled (5F10R probe)/SMN digoxigenin-labeled (132SE23 probe, specific to theSMN gene in the SMA locus), (II.) Glu 5–10 biotin-labeled (5F10R probe)/c41-cad digoxigenin-labeled (Φ-98-1 probe). The hybridization signals observed on 5p with the c41-cad probe are due to cross-hybridization with the genuine CDH12 gene.











