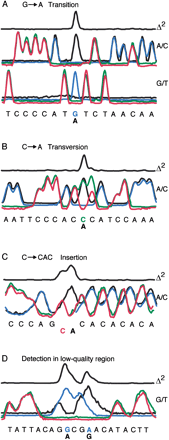
Representative traces of various polymorphisms. The Δ2 plot is generated by squaring the sum of the differences between the sample and reference traces for all four comparisons. (A) Two companion traces for a G→A transition polymorphism. Transition events detected in one trace are confirmed in the companion trace. (B) A/C trace for an C→A transversion displays mismatched coincident peaks. (C) Example of a C→CAC insertion. Insertion results in a sample/reference frame shift, which is corrected by realigning the traces after the insertion. (D) Positive polymorphism identification in a “low-quality” sequence region. Single-base resolution is not required for accurate base variation detection.











