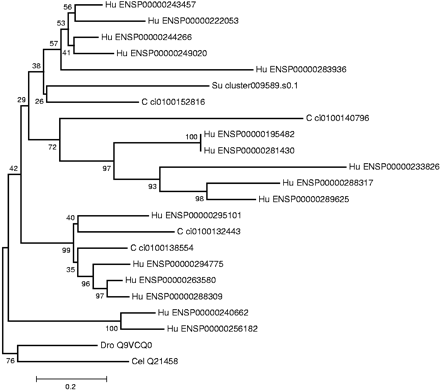
Example of a neighbor-joining phylogenetic tree generated for a CD-type gene family that includes a single sea urchin and multiple Ciona and human orthologs. This CD group represents a family of potassium channels. Most of the branches contain a Ciona ortholog, and hence most of the genes of this family were duplicated after the protostome/deuterostome split but before the vertebrate/chordate divergence as the CD/CDY groups include orthologs of genes that are simultaneously single copy in C. elegans, D. melanogaster and yeast (S. cerevisiae). Because the sea urchin sequence does not root the tree, the expansions might have taken place before the separation of the echinoderm lineage. Additional vertebrate-specific expansions like in the example above were repeatedly observed in the selected CD/CDY groups. The C. elegans and D. melanogaster node was used as an outgroup for the tree. Numbers at branch points are confidence values derived from 1000 bootstrap resamplings of the alignment data. The sequence distance is indicated at the bottom as substitutions per site. Human genes are abbreviated by Hu followed by the Ensembl gene identifier (release 4.28; see Methods), Ciona genes are abbreviated by C followed by the Ciona gene model identifier number (JGI Release1; see Methods), and the sea urchin sequence is abbreviated by Su followed by the sequence cluster identifier that can be retrieved from our database. Other abbreviations are (Dro) D. melanogaster and (Cel) C. elegans.











