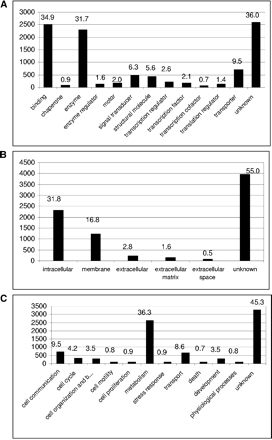
Distribution of the EST consensus sequences into the main Gene Ontology (GO) functional classes. Of 7146 consensus sequences, 5114 could be annotated in at least one of the three Gene Ontology (GO) defined functional classes. The numbers above the bars represent the percentage of all sea urchin clusters with a significant match to the SWISS-PROT database that are classified in the given functional class. The height of the bars and the ordinate give the number of proteins per group. (A) 4570 proteins could be associated with a molecular function, (B) 3219 proteins with a potential cellular component, and (C) 3910 proteins were associated with a specific biological process. A total of 2426 proteins were annotated in all three main categories.











