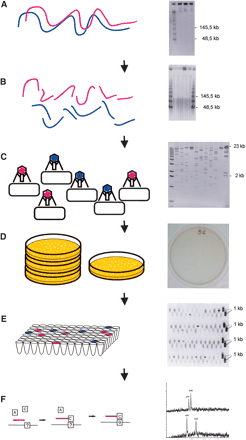
(Left) A schematic representation of the CSH method; (right) a corresponding example of “real data.” (A) Intact high-molecular-weight DNA is prepared, and the quality is checked by PFG electrophoresis. (B) DNA is sheared into pieces of ∼40–100 kb, as can be seen by PFG. (C) After end-repair, a cosmid/fosmid library is constructed by ligation of the fragments to a suitable vector, in vitro packaging, and transfection of bacteria. The quality of the library is checked by restriction analysis of a few randomly chosen clones. (D) The titer of the library is determined, and ∼10,000 clones are plated onto one plate. (E) The region of interest is amplified by PCR, and the positive pools are selected for subsequent analysis by the GOOD assay; as an example PCR3 of DNA11 is shown. (F) The genotypes of the SNPs are analyzed using the GOOD assay (Fig. 3).











