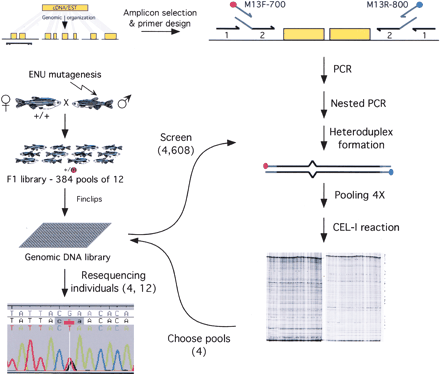
Efficient target-selected mutagenesis in zebrafish. Forty male zebrafish were mutagenized with ENU and outcrossed with wild-type females to generate a library of 4608 mutagenized F1 fish. Both males and females were finclipped and grouped in 384 pools of 12 fish per fish tank. DNA was isolated from the finclips and arrayed in twelve 384-well PCR plates. Amplicons of target genes were selected from local gene assemblies using GENOTRACE and primer picking software. Targets were amplified by PCR with gene-specific primers (1), followed by a nested PCR with internal gene-specific primers containing universal adaptor sequences (2), in combination with IR DYE-labeled universal M13 primers. Heteroduplexes were formed and samples were pooled fourfold. Pooled samples were incubated with CEL-I enzyme, and fragments were analyzed by denaturing polyacrylamide gel electrophoresis. Steps from PCR amplification to CEL-I incubation can be done in a completely unattended robotic setup. The four samples represented in a positive pool were reamplified from genomic DNA and subsequently resequenced. Fish carrying interesting mutations were recovered from pools of the F1 library by finclipping and resequencing each of the 12 fish of a pool.











