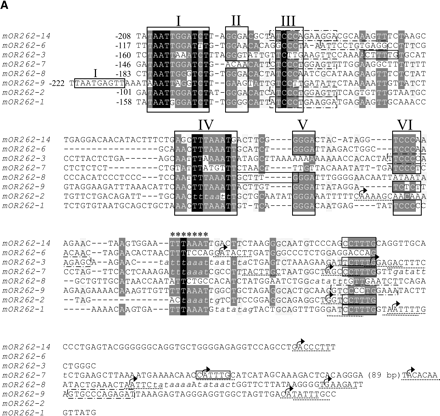
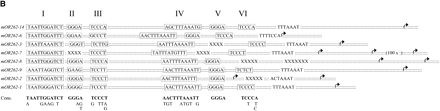
(A) Nucleotide sequence alignment of the conserved sequences in the 5′ regions of OR genes expressed in clustered neuron populations. The alignment was generated using T-COFFEE and manually edited. Identical nucleotide positions in all sequences are shaded in black, highly conserved positions are shaded in gray. Numbered frames (I-VI) indicate characteristic sequence motif blocks; the repeated motif block I found in mOR262-9 is also boxed. Stars indicate an additional AT-rich stretch located close to the most upstream TSSs (arrows). Putative initiator elements are underlined; TATA boxes and TATA-box-like sequence stretches are indicated in italics. Olf-1-like sequence motifs are framed by dotted lines, E-boxes and E-box-like sequences by solid lines. The numbers indicate the distance in bp to the nearest TSS. (B) Array of the conserved sequence motifs identified in the 5′ region of clustered OR genes. Nucleotides spacing the sequence motifs are indicated by the double dotted lines. An X is included at positions where no motif block could be identified in one particular sequence, to uphold the conserved distance of the remaining blocks. The resulting consensus sequence of the conserved motif blocks I-VI is given below.











