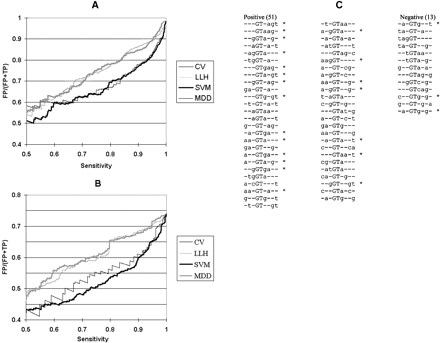
Three-way combinations of bases within the splice donor site. (A,B) False positive rate as a function of sensitivity in discriminating real and pseudo exon donor sites. Systematic variation of the threshold resulted in the different sensitivities. Classifying scores were from SVM (heavy black lines); multiple dependence decomposition (MDD, light black lines); consensus value (CV) calculated according to Shapiro and Senapathy (1987; heavy gray lines); and consensus values calculated by the log likelihood method (LLH, light gray lines). (A) The data set contained all of the real exons. Pseudo exons were defined as containing a simple GT as a potential donor site (no consensus value filter). (B) The data set contained all of the real exons. Pseudo exons were defined as having consensus values of at least 78. (C) Three-way combinations weighted most highly by SVM in distinguishing real from pseudo exons. The training set consisted of approximately 3400 real exons and 3200 pseudo exons, all of which exhibited donor site consensus values of at least 78. Positive and negative weights are listed separately, in descending weight order (absolute value). Asterisks denote agreements to the consensus. These 64 combinations allow SVM to perform at 92% of the accuracy achieved with the full set.











