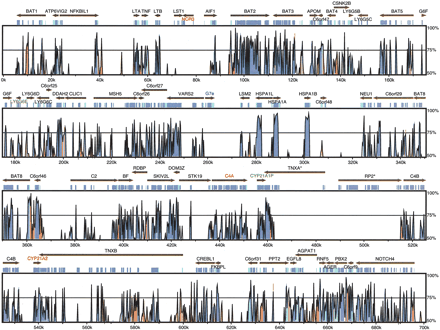
VISTA plot of the human and mouse MHC class III regions. Conserved sequences (percent identity >50%) are shown in different colors according to the type of their sequence: blue for coding regions, turquoise for UTRs, and red for CNSs. Two names with “*” (TNXA, RP2) denote gene fragments. Gene names are given for the human orthologs. Three gene names (NCR3, C4A, CYP21A2) are painted in red, to indicate their mouse orthologs are pseudogenes, and the names of two human pseudogenes (CYP21A1P and LY6G6E) are green. The two regions where the sequences do not align are due to a unique gene in mouse, G7e (name in blue), which resembles a viral envelope gene (Snoek et al. 1996) at 260 kb; and to several transposable elements that have inserted into the mouse genome between 470 and 510 kb. The approximate positions on the April 03 Goldenpath assemblies are chr6:31550009-32223670 (human) and chr17:33160937-33875007 (mouse).











