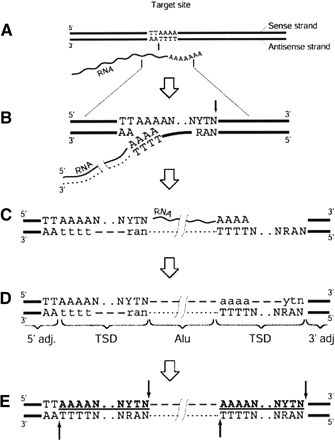
Model of Alu integration and generation of breakpoint near integrated Alu. This is based upon the B2, C2, and D4 rearrangements. (A) Enzymatic nicking in the presence of RNA indicated by vertical black arrow. (B) Synthesis of cDNA, indicated by dotted line, and formation of a second nick, indicated by black arrow on the opposite strand. (C) Completion of reverse transcription and DNA-dependent DNA synthesis, indicated by a dashed line and the lowercase letters, followed by ligation. (D) Elimination of RNA and synthesis of the second DNA strand. The integrated Alu element is surrounded by the target-site duplications (TSDs), usually 15-16-bp long. TSDs are marked in boldface and underlined. (E) Potential sites for secondary attacks by the L1 endonuclease, in 5′ and 3′ duplicated targets, are indicated by the black arrows. The 5′ flanking sequence contains an intact target TTAAAAN.NYTN; the 3′ duplicated target lacks the first two nucleotides (typically TT). Modified from Jurka (1997).











