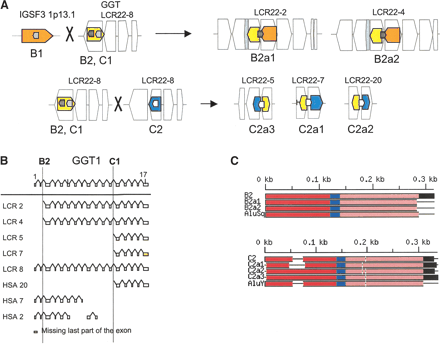
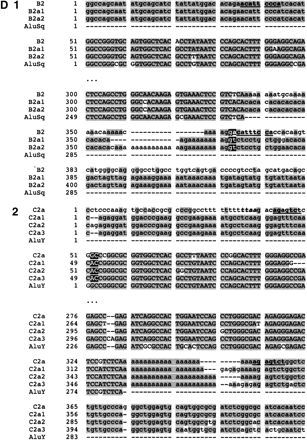
(A) Position of Alu elements involved in shaping GGT in the LCR22s. The GGT gene (yellow shapes; exonic orientation shown), IGSF3 (orange shapes), and DKFZp434p211 (blue shapes) within the block structure of the LCR22s are shown. Alu elements B1 (light gray), B2 (charcoal gray), C1 (light gray), and C2 (white) are illustrated in the “+” or “-” orientation. The duplicated copies of the Alu elements within their respective copies of GGT are illustrated. (B) Alus involved in breakpoints with respect to the genomic organization of GGT. The genomic organization of GGT was determined by comparing the cDNA sequence with the human genomic sequence by BLAST analysis. The position of Alus B1 and C1 with respect to the genomic organization of GGT is illustrated (dotted line). (C) Structure of Alu elements B2 and C2, including their duplicated copies. Each Alu monomer (red, pink) is illustrated on either side of the A-rich spacer (blue). The 3′ poly A tail is shown (black). Alu B2 and its copies are part of the Alu Sq subfamily (see Suppl. Fig. 4). Alu C2 and its copies are part of the Alu Y subfamily (see Suppl. Fig. 5). (D) Breakpoints within Alu targets. Alu repeats are shown in uppercase and flanking sequences in lowercase. The expected breakpoint positions are marked in red. Potential target site duplications are marked in boldfaced and underlined. (D1) Breakpoint within 3′ Alu target of Alu B2. The 5′ flanks of the Alu B2 substrate and its products B2a1, B2a2 are homologous, but this is not true of the 3′ flanks. The end of homology between Alu B2 and substrates corresponds to the position of the breakpoint. Alus B2a1 and B2a2 contain a variable (CA)n microsatellite within poly A tails, so it is difficult to mark the exact position. However, the presence of two Gs immediately flanking the poly A tail in both the substrate and products suggests the likely position of the breakpoint. During L1 endonuclease-mediated integration, the target sequence is duplicated at the 3′ end, except for the first two nucleotides. The position of the presumed breakpoint (GA) corresponds to start of the (partially preserved) 3′ Alu target-site duplication, i.e., 3′ duplicated target of the Alu insertion. Thus the recombination breakpoint coincides with the L1 endonuclease target site, which can be attacked by the L1 endonuclease. (D2) Breakpoint within 5′ target of Alu C2. The 3′ flanks of the Alu C2 and products C2a1, C2a2, C2a3 are homologous, but this is not true of the 5′ flanks; the end of homology between C2 and substrates, corresponds to the position of the breakpoint. The highlighted TTAA motif (yellow) corresponds to the putative original target; the first DNA nick probably occurred between TT and AA, 13 bp upstream of Alu C2a. The breakpoint within products is located 15 bp downstream of the expected first nick, perfectly fitting with the L1 endonuclease preference for second nick 15-16 bp downstream of the first one (Jurka 1997). Thus the breakpoint could be initiated by L1 endonuclease revisiting the original Alu target, and later repaired by homologous recombination. The second DNA nick probably occurred 2 bp downstream compared to the original insertion, and thus the first two nucleotides were not carried during the recombination event. See more details in Figure 9.











