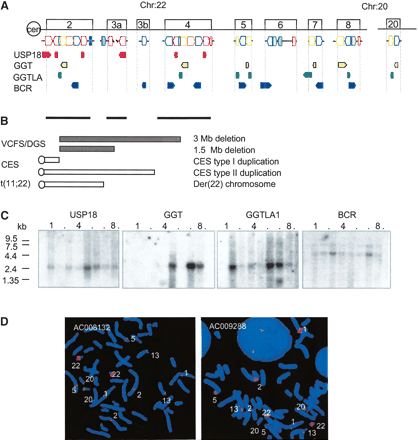
(A) Genes in the LCR22s. Four functional genes, USP18 (red), GGT (yellow), GGTLA (green), and BCR (blue) map to LCR22-2, LCR22-8, LCR22-7 and LCR22-6, respectively. Each has become copied during evolution, resulting in a complex pattern within blocks comprising LCR22s (colored blocks corresponding to LCR22 genes, orientation shown). The orientation of the genes and pseudogene copies with respect to the centromere is indicated. (B) Chromosome rearrangement disorders on 22q11.2. The bars under LCR22-2, LCR22-3a, and LCR22-4 depict the intervals harboring the common deletion endpoints, duplication endpoints, and translocation breakpoints in patients with VCFS/DGS, CES, and der(22) syndrome, respectively. (C) Northern blot analysis. We performed a Northern blot analysis using expression sequence tag (EST) DNA probes for USP18, GGT, GGTLA, and BCR. Autoradiograms of human multitissue Northern blots (Clontech) containing heart, brain (whole), placenta, lung, liver, skeletal muscle, kidney, and pancreas tissues were probed with radiolabeled PCR products from ESTs. The USP18, GGT, and BCR probes are derived from the last exon (except for USP18 in LCR22-3a), which would recognize all of the duplicated copies of each on chromosome 22 if transcribed. The band sizes for BCR are 4.79 kb, 7.50 kb; the expected sizes were 2.6 kb and 4.7 kb. The band sizes for GGT are 1.29 kb and 3.00 kb; the expected sizes were 1.8 kb and 2.5 kb. The band sizes for GGTLA are 1.58 kb and 2.8 kb; the expected size was 2.4 kb. The band size for USP18 is 1.95 kb; the expected size was 1.8 kb. (D) Low-stringency FISH mapping. Probes from LCR22-2 (GenBank AC008132) and LCR22-4 (GenBank AC009288) were used for low-stringency FISH mapping. Hybridization signals were detected in the vicinity of chromosomes 1p13, 2p11, 5p13, 13p11, and 20p12. The strongest signals were detected on chromosome 22q11, due to the presence of multiple copies of sequences contained within the LCR22 clones.











