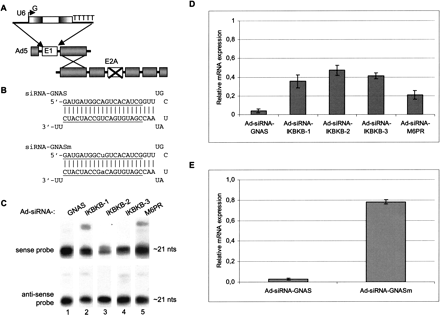
Adenoviruses efficiently deliver and express siRNAs with multiple target sequences. (A) Schematic representation of the U6-promoter-based siRNA expression cassette integrated in the adenoviral vector. The starting G and the stretch of Ts present for efficient transcription purposes are indicated. The replication-competent adenoviral molecule based on serotype 5 (Ad5), lacking the E1 and the E2A regions, is generated in Per.C6/E2A packaging cells by homologous recombination. Not drawn to scale. (B) Predicted secondary structures of hairpin RNAs derived from the adenoviral U6-driven expression constructs. The structure with GNAS (guanine nucleotide-binding protein α stimulating activity polypeptide 1) target sequence CGATGTGACTGCCATCATC (corresponding to the lower strand in the hairpin structure) is given as example, as well as that of the mutant version GNASm with a point mutation at the central position (lower case) of the target sequence (CGATGTGACaGCCATCATC). The variable sequence in the hairpin RNA that is dependent on the target sequence is underlined. (C) Expression analysis of small RNAs by Northern blotting using denaturing 15% polyacrylamide gels. Samples were isolated 3 d postinfection from U2OS cells that were infected at m.o.i. 3000 with the various adenoviral constructs as indicated. The blots were hybridized with labeled oligonucleotides recognizing either the antisense strand of the target sequence (upper panel, sense probe) or sense strand of the target sequence (lower panel, antisense probe). (D) Real-time reverse transcriptase (RT)-PCR expression analyses measuring the endogenous mRNA levels for GNAS, IKBKB (IKKβ, inhibitor of κ light polypeptide gene enhancer in B-cells, kinase β), or M6PR (mannose-6-phosphate receptor) of samples derived from U2OS cells 3 d after infection at m.o.i. 3000 with the indicated siRNA expression viruses. Values are plotted relative to samples derived from cells infected with control virus Ad-siRNA-GL2 and are normalized to GAPD (GAPDH, glyceraldehyde-3-phosphate dehydrogenase) mRNA as an internal reference. Values are obtained from three independent infection experiments; error bars show standard deviation. (E) Comparison of Ad-siRNA-GNAS versus Ad-siRNA-GNASm vectors by real-time RT-PCR analyses on samples derived from U2OS cells infected at m.o.i. 3000 6 d postinfection. See Figure 1B for sequences. Shown are the representative results from two independent experiments. Error bars indicate high-low values.











