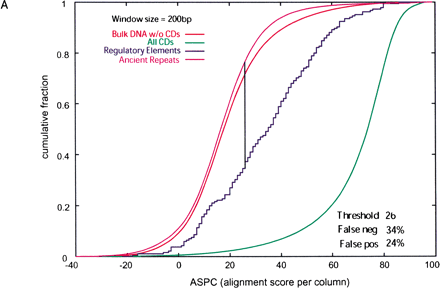
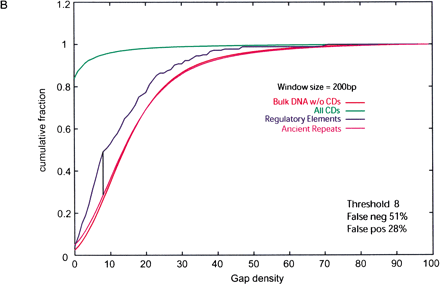
(A) Cumulative distributions of ASPC (alignment score per column) in 200-bp nonoverlapping windows from regulatory elements, ancient repeats, coding exons (cds), and bulk DNA alignments. The ASPC is calculated using the BLASTZ scoring scheme with a penalty for gaps. The vertical line represents the ASPC value at which regulatory element and ancient repeat distributions intersect (i.e., maximal distance between cumulative distributions). With this as a threshold, one obtains a certain percentage of false positives (ancient repeats above the threshold) and false negatives (regulatory elements above the threshold). (B) Cumulative distributions of gap density in 200-bp nonoverlapping windows from regulatory elements, ancient repeats, coding exons (cds), and bulk DNA alignments. The vertical line and percentages of false positives and false negatives are obtained as for ASPC in A, except that here false positives are ancient repeats below the threshold, and false negatives are regulatory elements above it.











