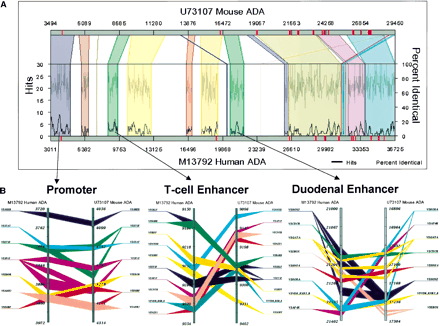
(A,B) Regulogram depiction of sharedcis-elements between two sequences in the context of their sequence similarity. The two sequences are represented as horizontal bars. The colored segments on these bars are exons. The regions of alignment are represented as different colored quadrilaterals that relate one sequence to another. Within each shaded block, the percent sequence similarity and the number of TF-binding sites are represented as two separate line graphs. The percent similarity is the average sequence conservation as determined by the BLASTZalgorithm and the shared cis-element hits are determined by an algorithm that uses a 200-bp moving window that looks through thecis-elements that are present within the conserved sequence block. Numbers are nucleotide positions. Regulogram can be clicked to zoom in or view the TF-binding sites that are in common between the two sequences at the click-point coordinate.











