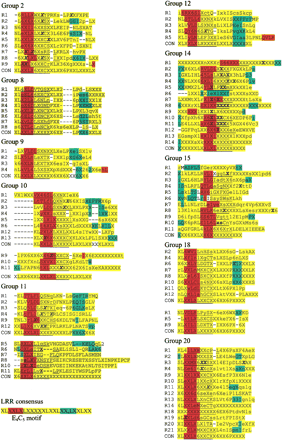
Secondary-structure predictions for LRRs from four groups with the highest number of positively selected sites. R# on left indicates the ordinal number for each LRR, and the sequence within each LRR represents the consensus from the alignment. Secondary-structure predictions are colored (yellow, coil; red, β-sheet; and blue, α-helix). The positions in which positive selection was detected are in bold and the sites that are part of the E4C5motif are underlined. CON is the consensus sequence among LRRs for each group. Amino acids represented in uppercase are invariant among sequences. Notation for variable amino acid sites are as follows: X, any amino acid; 1, D,N; 2, E,Q; 3, S,T; 4, K,R; 5, F,Y,W; and 6, L,I,V,M. Dashes are gaps introduced to better align consensus LRRs and do not necessarily represent gaps in the original sequence alignments. In group 11, repeats 5, 6, 7, 8, 10, and 11 were defined by their predicted secondary structure. Similarly, repeats 1, 5, and 9 of group 18 were defined by predicted secondary structure. For other groups, LRRs were defined either by Pfam or by their similarity to characterized R genes.











