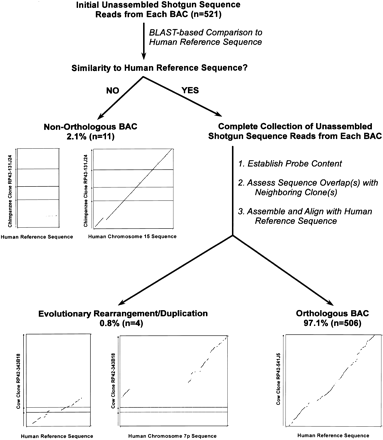
Process for validating orthology of bacterial artificial chromosomes (BACs) isolated with universal overgo hybridization probes. Initial unassembled shotgun sequence data (typically 96–384 sequence reads) generated from each BAC within a sequence-tiling path (see Fig. 3) are compared by BLAST to the corresponding human reference sequence. Each BLAST output is evaluated manually to determine whether or not there is significant similarity between the BAC and the human reference sequence (i.e., evidence of multiple independent alignments in the expected locations within the reference sequence). Clones with no significant sequence similarity are classified as nonorthologous, and this classification is confirmed by detecting similarity with another region of the human genome. For illustrative purposes, the results of analyzing the assembled sequence of a nonorthologous clone are shown (middle left). Specifically, dot-plot alignments (generated usingPipMaker) are shown between the sequence of chimpanzee BAC RP43–131J24 (GenBank AC087736; 172,676 bp) and the nonorthologous human reference sequence from chromosome 7q31 (80,001 bp) as well as the orthologous human chromosome 15 sequence (186,001 bp). For each BAC initially showing significant similarity with the human reference sequence, the complete collection of unassembled shotgun sequence reads are subjected to the following steps: (1) establish the presence of the expected overgo probes (i.e., those used during clone isolation and mapping); (2) assess that the appropriate sequence overlap(s) exist with any available neighboring clone(s); and (3) assemble the BAC sequence and align it with the human reference sequence. From these analyses, clones are either classified as being orthologous (bottom right) or containing an evolutionary rearrangement and/or a duplication (bottom left). As an illustration of the latter classification, dot-plot alignments are shown between the sequence of cow BAC RP42–343B18 (GenBank AC110663; 168,542 bp) and the human reference sequence from chromosome 7q21 (70,001 bp) as well as another sequence from chromosome 7p12 (260,001 bp). These alignments reveal that separate portions of this clone are orthologous to two distinct regions on human chromosome 7, indicating that the BAC spans an evolutionary rearrangement. In analyzing 521 isolated BACs by the steps depicted in this figure, 506 clones (97.1%) were found to be orthologous to the expected human reference sequence. To illustrate the typical results obtained with an orthologous clone, a dot-plot alignment is shown between the sequence of cow BAC RP42–541J5 (GenBankAC109796; 126,126 bp) and the expected human reference sequence (120,001 bp). Note that the dot-plots in this figure are not drawn to scale.











