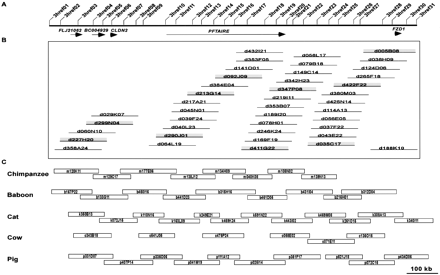
Orthologous sequence-ready bacterial artificial chromosome (BAC) contig maps constructed in multiple species. (A) Comparative BAC-based physical mapping was performed on a ∼1.2-Mb interval of human chromosome 7q21 containing the indicated five genes. In this region, 31 universal overgo hybridization probes were designed from human-mouse sequence alignments, with their locations shown to scale. The set of universal probes were used to screen six mammalian BAC libraries, with both probe-content maps and restriction enzyme digest-based fingerprint maps then deduced with the isolated clones. (B) The complete dog BAC contig map constructed for this region is depicted (but not drawn to scale). From this contig map, a minimally overlapping set of BACs (i.e., a sequence-tiling path) was selected (indicated by the shaded clones). (C) The resulting sequence-tiling paths selected from the chimpanzee, baboon, cat, cow, and pig BAC contig maps are shown, with the BACs drawn to scale based on the estimated clone overlaps and sizes.











