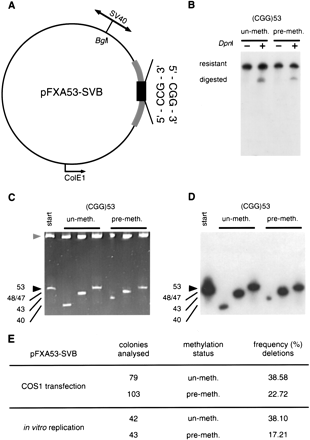
Effect of methylation on the genetic stability of (CGG)n repeats in a primate system. (A) The replication origin-containingSphI-HindIII fragment of the SV40 virus was cloned into the BglI site of pFXA53 (Table 1). The circular plasmid is drawn to scale. The location of the SV40-ori relative to the CGG tract defined the direction of mammalian replication. (B) The similar efficiency of replication of premethylated (SssI) and unmethylated templates in transfected COS1 cells (48 hr) or in vitro was confirmed by DpnI-resistance. Shown is a Southern blot of post-transfection KpnI/SacI digested DNAs hybridized with a 32P-labeled (CGG)10 oligonucleotide. (C) Replication products were analyzed for their repeat length alterations. Transfected or in vitro replication products were digested with DpnI (only DNAs that are products of mammalian replication are resistant to DpnI).DpnI-resistant replication products were transformed intoE. coli and DNAs were prepared from individual bacterial colonies, each derived from an individual product of mammalian replication. DNAs were analyzed for repeat changes by acrylamide gel electrophoresis. (D) Changes were confirmed to be the result of alterations in repeat numbers by electrotransfer and hybridization with a 32P-labeled (CGG)10 oligonucleotide. Shown is a representative gel with three single-colony/individual replication products of methylated (+) and unmethylated (−) templates. Individual products were also analyzed by DNA sequencing. (E) Frequencies of deletion events were scored. Background was subtracted from the mammalian mutation frequencies essentially as described (Panigrahi et al. 2002) (see Methods for details). Statistical analysis revealed a significant difference between the stability of methylated and unmethylated templates either in living cells (P = .014, χ2) or in vitro (P = .00088, χ2). These results are representative of at least two independent experiments.











