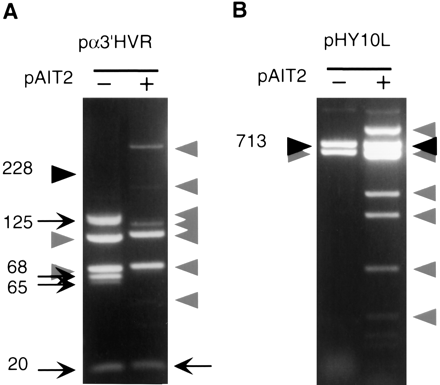
Effect of methylation on the genetic stability of minisatellite and satellite 3 repeats. Plasmids pα3′HVR and pHY10L containing the minisatellite 3′ of the α1-globin gene and the DYZ1 satellite 3 repeat, respectively (Table 1), were propagated for three subculturings in E. coli in the absence or presence of the SssI methylase-expressing plasmid pAIT2. Cells were harvested and plasmids isolated, restriction digested, and analyzed by 0.7% agarose gel electrophoresis and ethidium bromide staining. The trend through the three subculturings is reflected by the final subculture; hence only the third subculturing is shown for each plasmid. (A) pα3′HVR digested with BamHI/HindIII/ScaI is suitable to test for repeat length changes as it liberates the minisatellite-containing fragments, each of which are resolvable from vector and pAIT2 and vector restriction fragments (indicated by shaded arrowheads). The starting length of the 228 repeats is indicated by a filled arrowhead; all other faster migrating DNAs are products of deletion events, indicated by arrows. (B) Plasmid pHY10L digested with SspI/HindIII is suitable to test for repeat length changes as it liberates the satellite-containing fragment, which is resolvable from pAIT2 and restriction fragments (indicated by shaded arrowheads). The starting length of the 713 repeats is indicated by a filled arrowhead. The sister clone pHY10R, which is identical but cloned in the opposite orientation, yielded identical results (not shown). Gels in panels A and Bare representative of five or more independent experiments.











