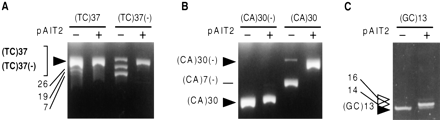
Effect of methylation on the genetic stability of dinucleotide repeats. Plasmids containing the indicated dinucleotides (TC)37, (CA)30, and (GC)13, cloned in both orientations; plasmids (Table 1) were propagated in E. coli for three subculturings (each representing 25 generations) in the absence or presence of theSssI methylase plasmid. Cells were harvested and plasmids isolated, restriction digested, and analyzed by PAGE and ethidium bromide staining. The trend through the three subculturings is reflected by the final subculture; hence only the third subculturing is shown for each plasmid. The starting length of each repeat is indicated by filled arrowheads, slower migrating repeat expansion products are indicated by open arrowheads, and all other faster migrating DNAs are products of deletion events. (GC)n expansions are indicated by open arrowheads. For each experiment, repeat length changes of individual expansion and deletion products were confirmed by DNA sequencing. (A) (TC)37 and (TC)37(−) digested withBamHI/NdeI analyzed on 8% PAGE. (B) (CA)30 and (CA)30(−) digested with BamHI/HindIII analyzed on 8% PAGE. (Because of the opposite cloning orientation and the polylinker, the (CA)30 restriction fragments migrate faster than their sister (CA)30(−) restriction fragments. (C) (GC)13 digested with BamHI/HindIII analyzed on 20% PAGE. Gels in panels A, B, and C are representative of five or more independent experiments.











