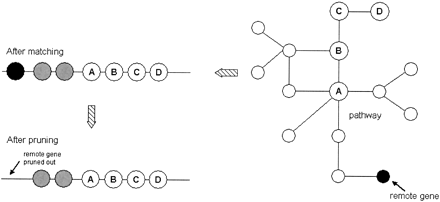
Graphical illustration of pruning procedure. Nodes with the labelsA,B,C,D in the pathway graph and the genome (line) are matched enzymes. The black vertex is the remote gene, which is three reaction steps away from the nearest gene (A) in the graph and three open reading frames (ORFs) away from the nearest gene (A) on the chromosome (gray genes are genes that were not matched). Consequently, the black vertex gets pruned from the cluster. The idea of pruning is implemented by computing the shortest distance in the graph from each matched vertex to the nearest matched vertex. A special case occurs when only two genes are reported as a possible operon. If their metabolic distance is equal to 3, they are pruned out.











