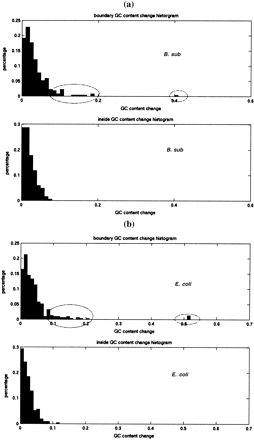
GC content change inside operons and at operon boundaries. Histogram of GC content change (x-axis, 0 ∼1) in operon and boundary regions inE. coli (a) and B. subtilis (b). GC content change was computed from two genes next to each other. If both of them are inside an operon, it is counted as inside operons. If either one is outside of the operon, it is counted as at the boundaries. GC content change is calculated for each gene. Ellipses mark the high GC transitions at the boundaries of the operons.











