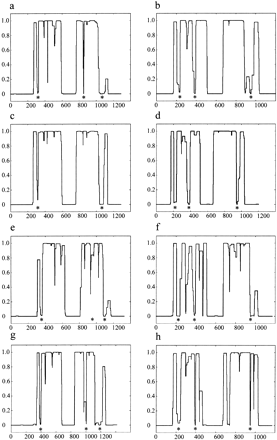
COILS prediction of SMC2 and SMC4 orthologs. TheCOILS output for Homo sapiens SMC4 (a),H. sapiens SMC2 (b), Xenopus laevi SMC4 (c), X. laevi SMC2 (d), fission yeast (Schizosaccharomyces pombe) SMC4 (e), fission yeast SMC2 (f), budding yeast (Saccharomyces cerevisiae) SMC4 (g), and budding yeast SMC2 (h). The vertical axis denotes the probability of the amino acid sequence adopting a coiled-coil structure based on a scanning window of 28 residues. Conserved disruptions in the coiled-coil structure prediction to the condensin SMC sequences are indicated beneath the COILSprofile with an asterisk.











