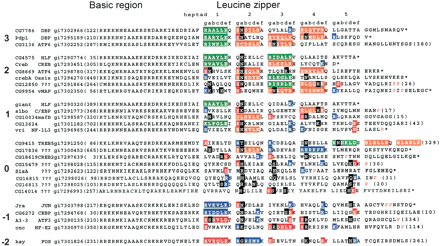
Alignment of 27 identified Drosophila melanogaster B-ZIP motifs using the single letter amino acid code. The proteins are arranged into groups based on the number of attractive g↔e‘interactions minus repulsive g↔e‘ interactions ranging from 3 to –2 pairs. The column starts with the name of the protein. Next is the name of the closest mammalian homolog, followed by the gi# for the Drosophila melanogaster sequence. The protein sequence of the B-ZIP motif follows. The number of amino acids from the predicted N terminus of the protein to the B-ZIP motif is given in parentheses. The C terminus of each sequence is either the natural C terminus denoted by an asterisk (*), or a truncation with the number of amino acids to the C terminus in parentheses. To help visualize the potential g↔e‘ interactions, we grouped heptads (gabcdef). If both the ‘g’ and ‘e’ positions contain charged amino acids, we color both of these amino acids and the intervening ones (gabcde). We use green for the attractive basic–acidic pairs (R↔E and K↔E, K↔D), orange for the attractive acidic–basic pairs (E↔R, E↔K, D↔K, and D↔R), red for the repulsive acidic pairs (E↔E, D↔E, and E↔D), and blue for the repulsive basic pairs (K↔K, R↔K, K↔R, and R↔R). If only one of the two amino acids in theg↔e‘ pair is charged, we color only that amino acid: red if it is acidic and blue if it is basic. If the ‘a’ or ‘d’ positions contain polar or charged amino acids, they are colored black. The α-helix breaking prolines, indicative of the C terminus of the leucine zipper, are colored red.











