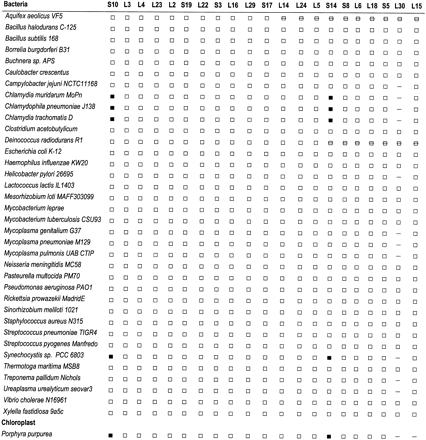
Unique shared-derived characters of the ribosomal super operon that unite cyanobacteria and Chlamydiaceae. Two unique shared-derived characters on the ribosomal super operon (the loss of ribosomal proteins S10 and S14) unite the Chlamydiaceae and cyanobacteria to the exclusion of other bacteria with genomes that have been completely sequenced (black boxes; note that S10 and S14 are present elsewhere on the chromosome). Loss of L30 (dashes; note that L30 does not appear to be present elsewhere in these genomes, according toTBLASTN analysis) is not a unique shared-derived character to the exclusion of all other bacteria but offers further support for a relationship between the Chlamydiaceae and cyanobacteria. In addition, all 10 chloroplast genomes examined (Porphyra purpurea chloroplast is shown as a representative) and an unfinished cyanobacterial genome (Synechococcus spp.) also share the same characters (i.e., loss of S10, S14, and L30 from the super operon); however, the chloroplasts are missing additional genes from this region (i.e., L15 in the region shown) that have been primarily transferred to the plant nucleus. Boxes with strikethroughs mark genes that have relocated in Deinococcus andAquifex to form a separate operon. Note that the genome annotation for Aquifex did not report L29; however, we did positively identify this gene in Aquifex usingTBLASTN. Another unique character uniting Chlamydiaceae, cyanobacteria, and the chloroplast, which is not illustrated in this figure, is that S10 is found as part of the separate S7/S12 operon in only the Chlamydiaceae, cyanobacteria, and chloroplast sequences examined.











