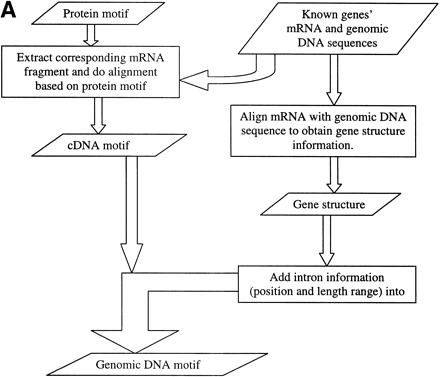
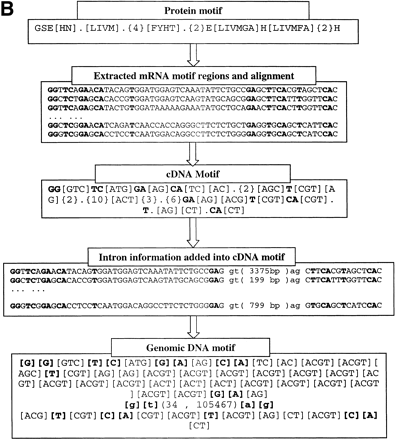
(A) Method to construct genomic DNA motif. Three steps were taken to construct the genomic DNA motif for a given family from known protein, mRNA, and genomic DNA sequences of the family. First, based on the locations of the protein motif in protein sequences, the corresponding mRNA regions were extracted and aligned to reveal the consensus pattern. Each site in the consensus pattern would include all nucleotides existing in the mRNAs at the site. Second, gene structures were obtained by aligning mRNA with genomic DNA sequences, and the intron information was collected. Third, the intron information was incorporated into the cDNA consensus pattern to generate the final genomic DNA motifs. (B) Motif construction example in the CA family. Conservative sites in DNA motifs are in bold font. Donors and acceptors of introns are in small letters. The number in the brackets in the DNA sequence alignments is the intron length in each gene. In the final Genomic DNA Motif, the two numbers separated by a comma in the parentheses (34 , 105467) are the minimum and maximum lengths of the intron in this position. Each pair of brackets in the DNA motif represents one site in the sequence, and the bases within each pair of brackets represent all possible nucleotides at that site.











