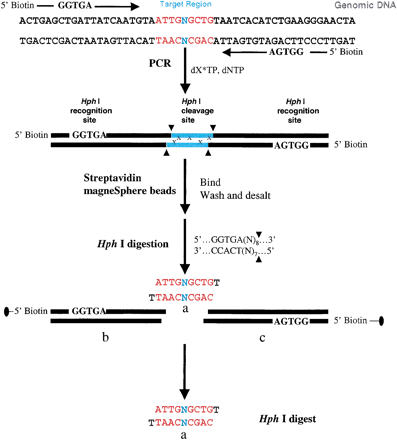
A schematic representation of our strategy for PCR amplification of the target region of the uncalled base in chromosome 16. The synthetic PCR primers are biotinylated and the restriction recognition sites ofHph I are included at the 5′ end of each primer. HphI has the recognition sequence of 5′…GGTGA (N8)…3′ and 3′…CCACT (N7)…5′, which dictates the restriction cleavage at the 8th base from the 5′ end and the 7th from the 3′ end, resulting in a protruding base at the 3′ end of each strand of the target region. The nucleoside, N, representing the complimentary sequences of the genomic DNA is included in the primer sequences. After PCR amplification and labeling with13C/15N-labeled nucleoside, x, streptavidin MagneSphere paramagnetic beads are added to the solution to capture the biotinylated PCR products through biotin-streptavidin interactions. The PCR products linked to the beads are washed and then subjected to theHph I restriction digestion, and the target sequence-containing SNP site(s), “Hph I mid-digest,” or a, is released in solution. The biotin-labeled end of the digest portions (Hph I recognition sequence plus [N]8/[N]7 flanking regions), b and c, are attached to the paramagnetic beads and trapped with a MagneSphere Technology Magnetic Separation Stand. The labeled “Hph I mid-digest,” a, is then desalted via a nitrocellulose membrane and dried for MALDI-TOF analysis. In MALDI-TOF spectra, the 50%13C/15N-labeled nucleoside, x, will induce a mass shift in the product, a, depending on the number of labeled nucleosides in the target fragment. From the molecular mass of the unlabeled product and the number of peaks present in the labeled product, the number of labeled nucleosides incorporated in the amplified product is determined.











