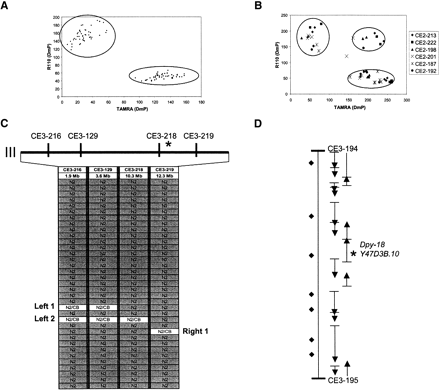
Sample fluorescence polarization-template directed incorporation (FP-TDI) mapping data. (A) High quality discrimination data from the FP-TDI assay of 48 random SNP markers on control genomic DNA from N2 and CB4856 strains. SNPs are detected through analysis of a single base pair extension from a sequencing primer that hybridizes just adjacent to the polymorphic nucleotide (Chen et al. 1999). Clusters are readily identified and unambiguous. (B) FP-TDI data from eight recombinant samples (crude worm lysates), each tested with six SNP markers. In this case, the FP-TDI kit used distinguishes A from T, the most prevalent SNP change in C. elegans. (C) FP-TDI analysis with the four Tier 2 SNP markers on chromosome III using 35 samples from a mapping cross to localizedpy-18. We localized the recessive gene to a 2.0-Mb subregion of chromosome III. SNP data are converted to table format for interpretation, and the informative recombinants that defined the interval are labeled Left 1, Left 2, and Right 1. (D) The 97-kb interval defined by the Tier 3 markers CE3–194 and CE3–195 is shown. There are 16 predicted genes in this region (AceDB version WS-48); for Tier 4 mapping there are nine predicted substitution SNPs in this interval. RNAi of the predicted gene sequenceY47D3B.10 is known to give a Dumpy (Dpy) visible phenotype (Hill et al. 2000), indicating that this gene is a strong candidate fordpy-18. We confirmed by directed sequencing thatdpy-18(e364) did contain a mutation G to A in exon 3 altering the TGG(Trp) codon to TAG(Stop).











