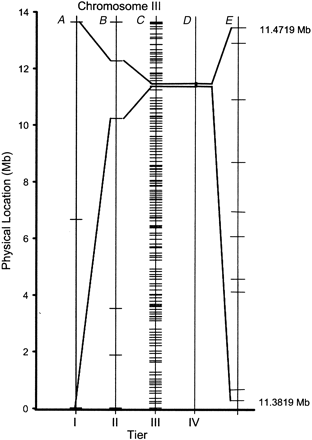
Tiered mapping strategy. Shown is a schematized mapping workflow. (A) Tier 1 mapping localizes the gene of interest to a chromosome by assessing linkage to one centrally located SNP. (B) Tier 2 mapping localizes the gene to a subregion of the chromosome. This region can vary in size between 1 and 6.7 Mb. In thedpy-18 example shown, the gene falls between two SNP markers that are 2 Mb apart on chromosome III. This resolution is routinely achievable by genotyping 30–60 recombinants. (C) Tier 3 mapping begins by identification of informative animals with recombination breakpoints within the region defined by Tier 2 and then fine mapping with Tier 3 markers to narrow the region of interest. Fordpy-18, Tier 3 mapping localizes the gene to a region as small as 97 kb and thus narrowed the candidate region to ∼0.1% of the worm genome. The number of recombinants required to achieve this mapping resolution will depend on the local recombination rate (Fig. 1B; Genetic Map Position from Wormbase; Stein et al. 2001) along the chromosome in the vicinity of the mutated gene. (D andE) Tier 4 mapping and/or mutation detection. Further refinement of the candidate interval occurs by validation of additional SNP markers and genotyping of informative recombinants. Fordpy-18, the location of the predicted substitution SNPs located within the 97-kb region of chromosome III are shown. During this fine-mapping process, candidate gene approaches such as cosmid rescue or RNA interference (RNAi) can also be used to help identify the mutation.











