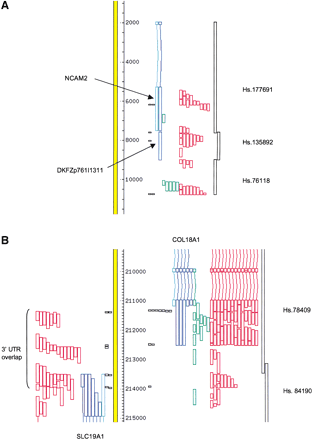
Examples of extended and overlapping 3′ untranslated region (UTR). Alignments of transcripts to the genome (yellow bar) were visualized using ACEDB. The direction of transcription is bottom totop on the left and top to bottomon the right. The direction of transcription of unspliced expressed sequence tags (ESTs) was based on their annotation, except for the unspliced ORESTES sequences, which were arbitrarily assigned to the right-hand strand. The orientation of 3′ tags was deduced from the polarity of the poly(A) tract. Light blue: RefSeq sequences; dark blue: full-length cDNA sequences; green: ORESTES sequences; and red: EST sequences. 3′ tags are represented by black boxes, with one box per cluster member. Regions covered by UniGene clusters are indicated on the right. (A) 3′ terminal exons of theNCAM2 gene. (B) Overlapping 3′ ends of theCOL18A1 and SCL19A1 genes. Many ESTs derived from the COL18A1 gene were omitted for clarity.











