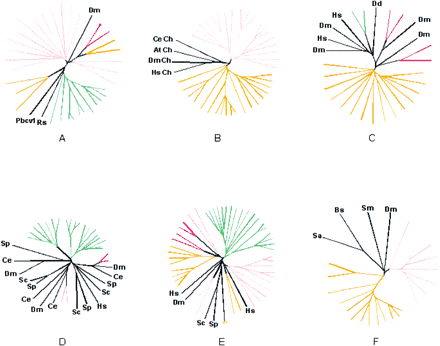
Phylogenetic analysis of selected eukaryotic lineage-specific expansions. Groups supported by a bootstrap value >70% are colored pink for Drosophila melanogaster, red for Homo sapiens, orange for Caenorhabditis elegans, green forArabidopsis thaliana, and yellow for Schizosaccharomyces pombe. (A) Prolyl hydroxylases. (B) Small molecule kinases (Ch stands for choline kinase). (C) Patched-like protein. (D) MAP-Kinases. (E) P450 family hydroxylases. (F) MBOAT membrane acyltransferases. (At)Arabidopsis thaliana; (Bs) Bacillus subtilis; (Ce)Caenorhabditis elegans; (Dd) Dictyostelium discoideum; (Dm) (Drosophila melanogaster; (Hs) Homo sapiens; (Pbcv1) Paramecium bursaria Chlorella virus 1; (Rs) Ralstonia solanacearum; (Sa) Staphylococcus aureus; (Sc) Saccharomyces cerevisiae; (Sm)Sinorhizobium meliloti; (Sp) Schizosaccharomyces pombe. Complete tree descriptions (full lists of GI numbers or gene names, and bootstrap values) are available in the Supplementary Material online atftp://ncbi.nlm.nih.gov/pub/aravind/expansions, andhttp://www.genome.org..











