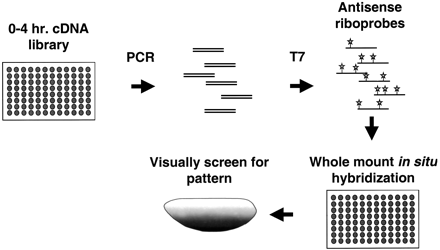
Profiling Patterned Transcripts in Drosophila. Bacterial cultures of individual clones from a 0 to 4 h cDNA library were used to seed PCR reactions driven by oligonucleotides that are complementary to sequences that flank the cDNA insertion site. The cDNA products were then purified on glass fiber filters in a 96-well format. This DNA was used to program transcription from a T7 promoter that resides just 3′ to the cDNA insert. Digoxigenin-UTP was included to label the antisense RNA probes produced. These probes were used subsequently in hybridizations to whole mount embryos in situ. The embryos were then visually screened using standard light microscopy to identify patterns of expression. All steps of the process were performed using a robotics workstation. The antisense RNA probes can be stored for extended periods of time and provide a reusable resource since only a small fraction of the probe is needed to subsequent steps. The slowest step in the process was the hybridization in situ, which took nearly 16 hours to complete. However, due to extended incubations, four plates could be staggered through the process simultaneously, yielding an effective throughput of 384 clones for each overnight run.











