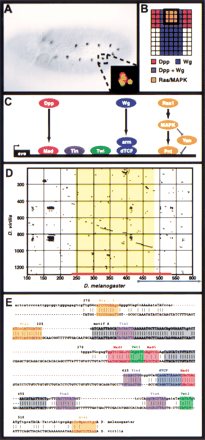
The Eve MHE provides a model for the transcriptional integration of multiple intercellular signals. (A) Stage 11Drosophila embryo stained with antibodies against Eve. A small cluster of cells in the dorsal mesoderm of each segment, the Eve-positive muscle and cardiac progenitors, express Eve. Anterior is to the left. Inset: Double-staining for Eve (green) and the MHE reporter construct (red) shows that the MHE is sufficient to drive expression in the Eve-positive cells (Halfon et al. 2000). (B) Signaling events required for Eve expression in the dorsal mesoderm. One hemisegment is represented, with dorsal to the top and anterior to the left. Expression of Eve is induced in cells that receive signaling from the Dpp, Wg, and Ras/MAPK pathways (Carmena et al. 1998). (C) The transcriptional code used at the MHE. The signal-responsive TFs, that is, Mad, dTcf, and Pnt bind along with the mesodermal selector proteins Twi and Tin to activate transcription. (D) Dot plot showing that the MHE sequence (x-axis, red bar) is conserved in D. virilis (y-axis). The area of extensive homology (on the diagonal) is shown in yellow; little homology exists flanking this region for several hundred base pairs. An alternate sequence recently proposed to have MHE-like activity (Knirr and Frasch 2001) is indicated by the blue arrow; note that most of this sequence lies in nonconserved regions. (E) Sequence alignment of MHE and vMHE. Known MHE binding sites are shown in color; gray boxes indicate conserved regions where functional sites have not yet been identified. Base pair numbering corresponds to that shown inD.











