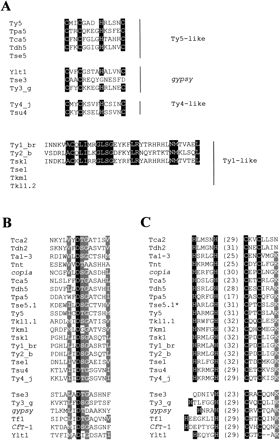
Amino acid sequence alignment of the conserved motifs in ORF1 and ORF2. (A) Sequence alignment of the RNA-binding domain located at the carboxy terminus of gag homologs in hemiascomycetous LTR-retrotransposons. The highly variable CX2CX4HX4H motif is only conserved in few hemiascomycetous retroelements including Ty3-like, Ty4-like and some Ty5-like elements. Ty1 and Ty2 lack the CX2CX4HX4H consensus motif but have a nucleic acid-binding motif homologous to a consensus prokaryotic DNA-binding sequence. Conserved residues are in white on black background. (B) Multiple sequence alignments of the protease active site. The universally conserved aspartic acid is shown in white on black background. The conservative substitutions found in more than 75% of the elements are in white on gray background. (C) Multiple sequence alignments of the zinc finger of integrase. Highly conserved residues of the HHCC zinc finger are shown in white on black background. The numbers within brackets indicate the number of residues constituting the loop between HH and CC. As the environment of the zinc finger is also conserved, the conserved K residue 4 amino acids downstream from the second C residue is shown in white on grey background. The star indicates that Tse5.1 and Tse5.2 possess exactly the same zinc finger, the sequence of the protease of Tse5.2 is not available.











