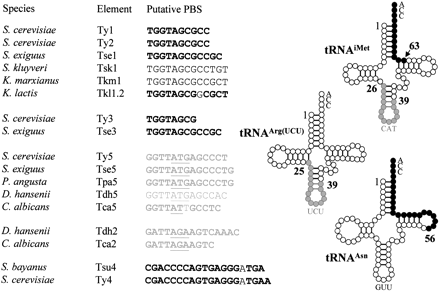
Primer-binding sites of LTR-retrotransposons and tRNA molecules of hemiascomycetous yeasts. The TGG trinucleotide starting most of the PBSs is complementary to the CCA trinucleotide present at the 3' end of all tRNAs and is characteristic of PBSs that utilize the 3' end of a tRNA as a primer. In contrast, the PBSs that are complementary to internal positions of tRNAs include the anticodon stem-loop (underlined). The tRNA molecule, residues that are complementary to PBS are indicated in black when located at the 3' end of the molecule and in gray when located in the internal part. Similarly, PBS sequences are written in black or gray depending on their positions. PBS sequences in bold indicate that the PBS was deduced from the comparison with the tRNA gene identified in the species. The mismatches in the PBS sequences of Tkl1.2, Tca5, Tsu4 and Ty4 are not in bold.











