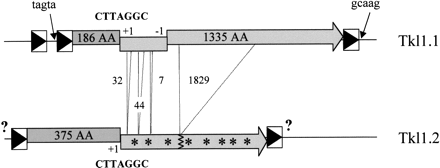
LTR retrotransposons of K. lactis. Two copies of the same element were identified in K. lactis, namely Tkl1.1 and Tkl1.2. The 5' and 3' LTR (boxes containing a black triangle) of Tkl1.1 only share 93 % nucleotide identity and the complete element is not flanked by the expected 5 bp nucleotide duplications of the target site. Sequences adjacent to Tkl1.1 LTRs are indicated by arrows. One of the two overlapping ORFs is homologous to gag (dark gray boxes containing the size of the protein), but lacks the first 189 aa at the N-terminus, and the other one is homologous to pol (light gray boxes containing the size of the protein) but has a one nucleotide deletion between positions 1909 and 1931, which generates a –1 frameshift. Tkl1.2, contains the complete gag gene, the consensus frameshift motif CTTAGGC indicated by “+1” found in Tkl1.1 and a second gene (light gray boxes containing stars) interrupted by numerous stop codons and small (7, 32, 44 nt) or large (1829 nt) deletions (dotted lines; sizes are in nt) compared to Tkl1.1. The sequences of the flanking LTRs have not been determined (question marks). Probes for specific regions of both copies (in the 5' part ofgag for Tkl1.2 and in the part of pol gene that is deleted in Tkl1.2 for Tkl1.1) were used in Southern hybridizations (data not shown) and confirmed that only one copy of each element is present in the K. lactis strain studied.











